InhibNet

InhibNet: Competitive Inhibition of Polymer Networks
1. Modulus Prediction
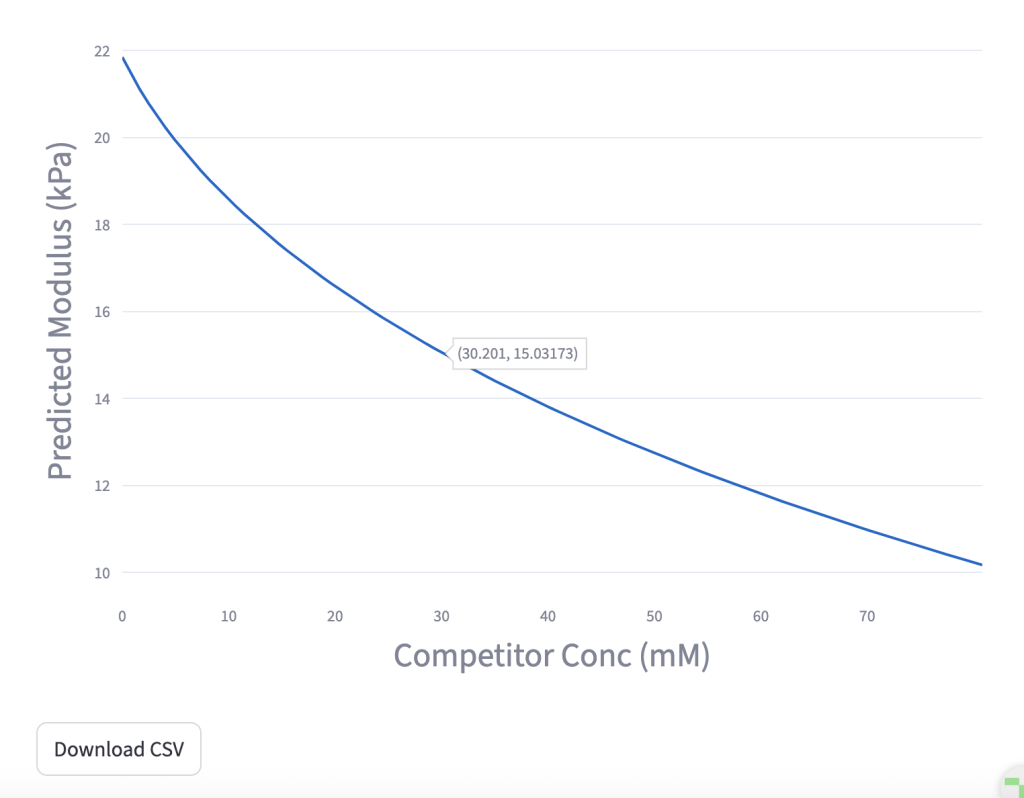
Predict the hydrogel stiffness (modulus) as a function of polymer concentration, crosslinking equilibrium constant, competitor equilibrium constant, and the concentrations of polymer and competitor in solution.
2. Ka prediction from modulus
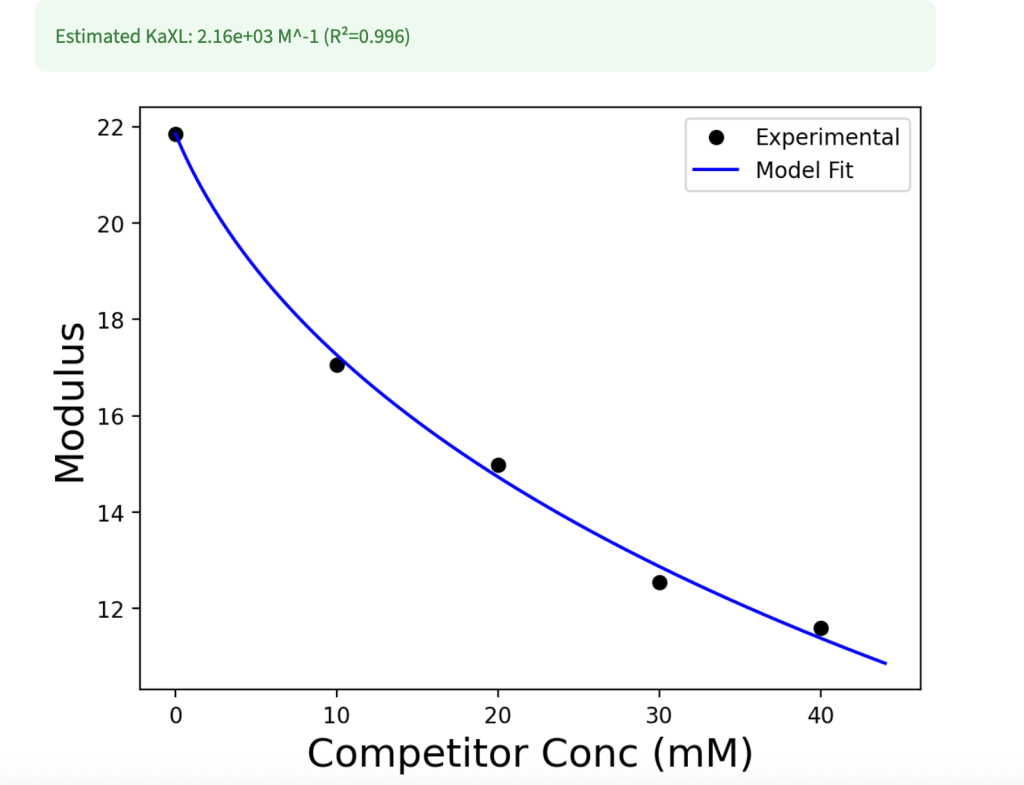
Here, we present a method to predict the equilibrium constant (Ka) of either the crosslinking interaction within a polymer network or a competing species. By inputting the competitor concentration and experimental modulus data, users can specify which Ka value is known and which should be determined through fitting.
3. Tau Fitting
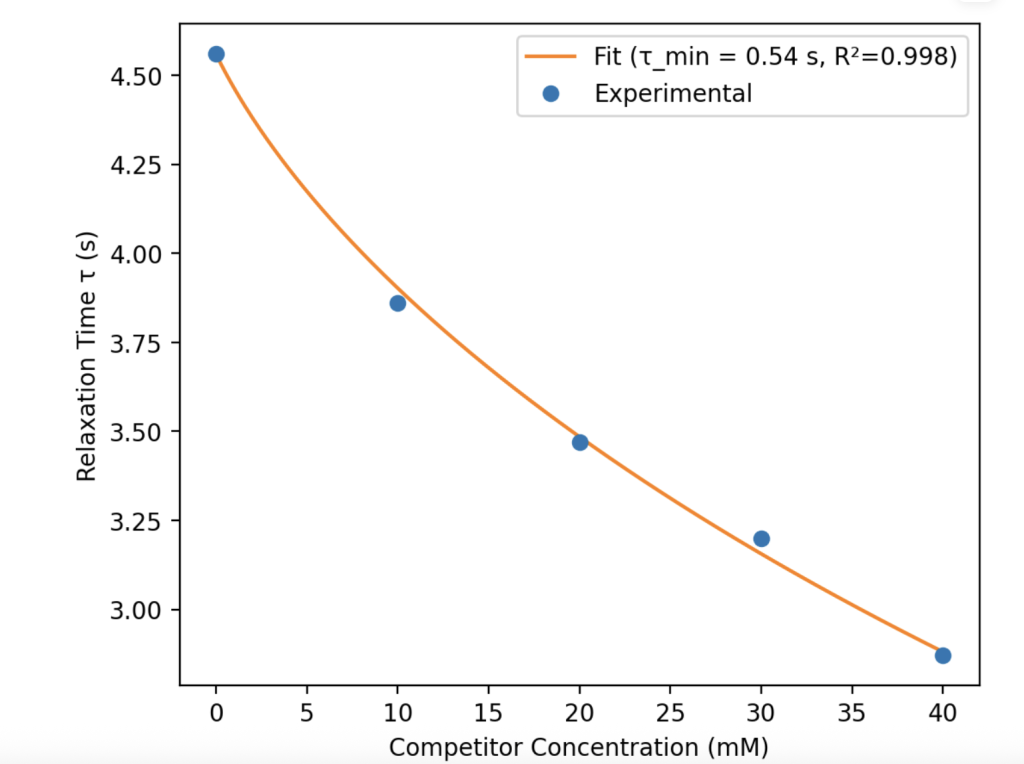
This module fits relaxation time (τ) values that change under competition using a Langmuir decay isotherm to capture how τ varies with competitor concentration.
4. 3D Surface Explorer
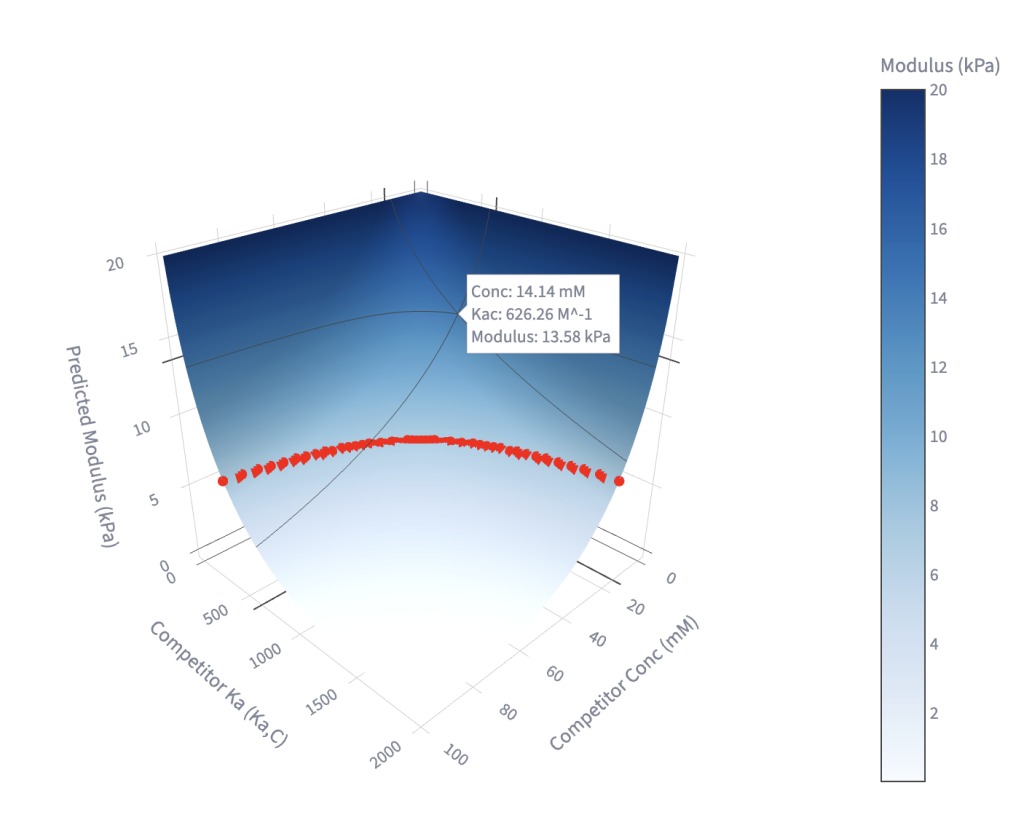
This tool generates an interactive 3D plot showing the relationship between competitor Ka, competitor concentration, and the resulting modulus. The crosslink Ka is held constant. Users can explore the range of modulus values achievable for a given competitor, or identify the competitor parameters required to reach a target modulus, highlighted by the red band.
5. Network Visualization
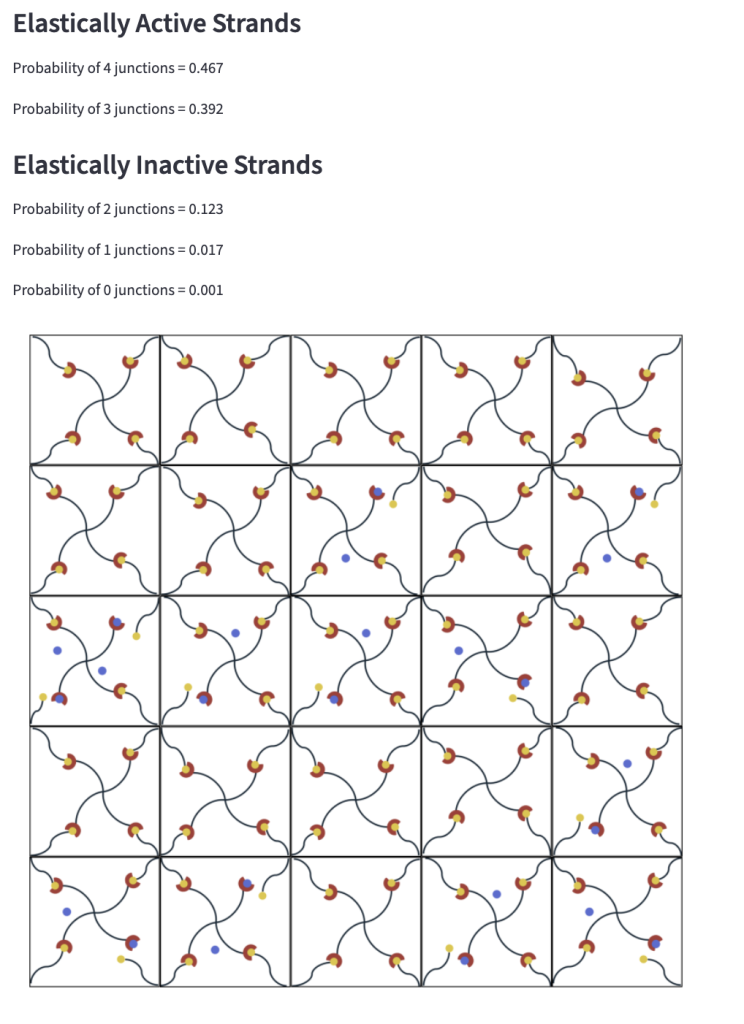
This tool generates a 2D map illustrating the predicted structure of a polymer network under varying associations of competitors and crosslinks. When no competitor is present, an alternative image set is used to represent the baseline state. The visualization demonstrates the mean-field approximation and enables quantification of the fraction of elastically active chains within the system.